4i
[4]:
import os
import pandas as pd
import numpy as np
import scanpy as sc
import squidpy as sq
import anndata as ad
import matplotlib.pyplot as plt
import seaborn as sns
[5]:
import pysodb
[6]:
sc.set_figure_params(vector_friendly=False,format='pdf',transparent=True,dpi=50)
plt.rcParams["figure.figsize"] = (8, 8)
sns.set_style('white')
load data using pysodb
[7]:
sodb = pysodb.SODB() # Initialization
/home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/urllib3-1.26.12-py3.9.egg/urllib3/connectionpool.py:1045: InsecureRequestWarning: Unverified HTTPS request is being made to host 'gene.ai.tencent.com'. Adding certificate verification is strongly advised. See: https://urllib3.readthedocs.io/en/1.26.x/advanced-usage.html#ssl-warnings
warnings.warn(
[8]:
# this dataset is from publication "Multiplexed protein maps link subcellular organization to cellular states"
[9]:
# link in SODB: https://gene.ai.tencent.com/SpatialOmics/dataset?datasetID=62
[10]:
adata = sodb.load_experiment('gut2018multiplexed','cell_129')
# the first parameter is the name of the dataset
# the second parameter is the name of one experiment in the dataset
load experiment[cell_129] in dataset[gut2018multiplexed] from /home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/pysodb-1.0.0-py3.9.egg/pysodb/cache/gut2018multiplexed/cell_129.h5ad
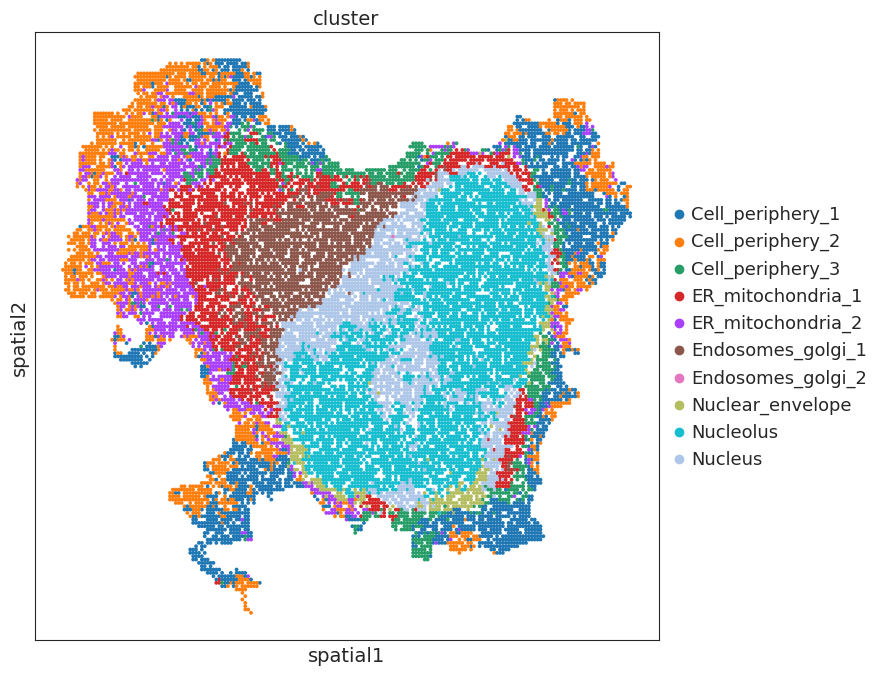
display clusters
[14]:
sc.pl.embedding(adata,basis='spatial',color='cluster',s=30)
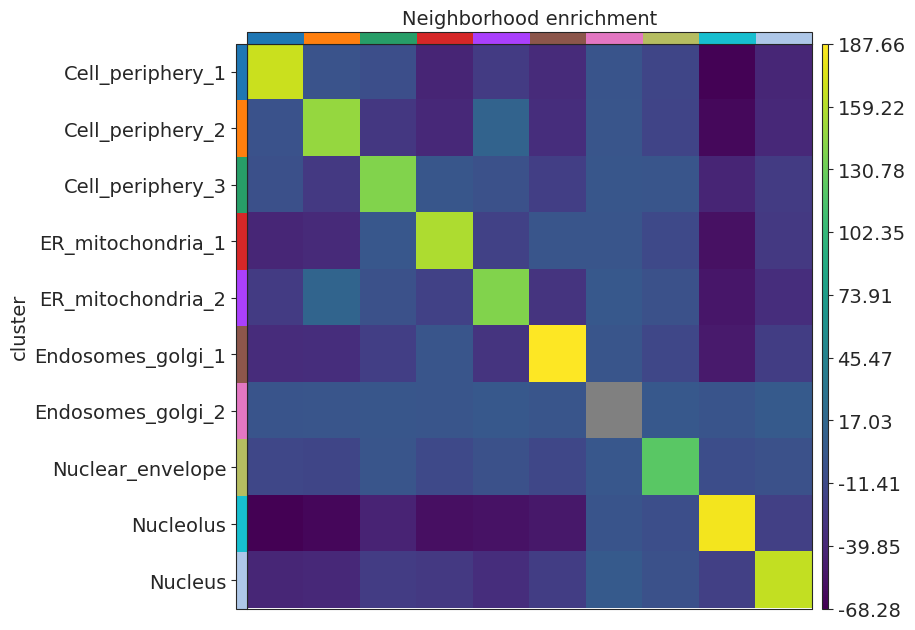
Neighborhood enrichment
[15]:
sq.gr.spatial_neighbors(adata)
sq.gr.nhood_enrichment(adata, cluster_key="cluster")
sq.pl.nhood_enrichment(adata, cluster_key="cluster")
/home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/tqdm-4.64.1-py3.9.egg/tqdm/auto.py:22: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1000/1000 [00:01<00:00, 982.36/s]
/home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/squidpy-1.1.2-py3.9.egg/squidpy/gr/_nhood.py:182: RuntimeWarning: invalid value encountered in divide
zscore = (count - perms.mean(axis=0)) / perms.std(axis=0)
/home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/squidpy-1.1.2-py3.9.egg/squidpy/pl/_graph.py:239: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
ad = AnnData(X=array, obs={cluster_key: pd.Categorical(adata.obs[cluster_key].cat.categories)})
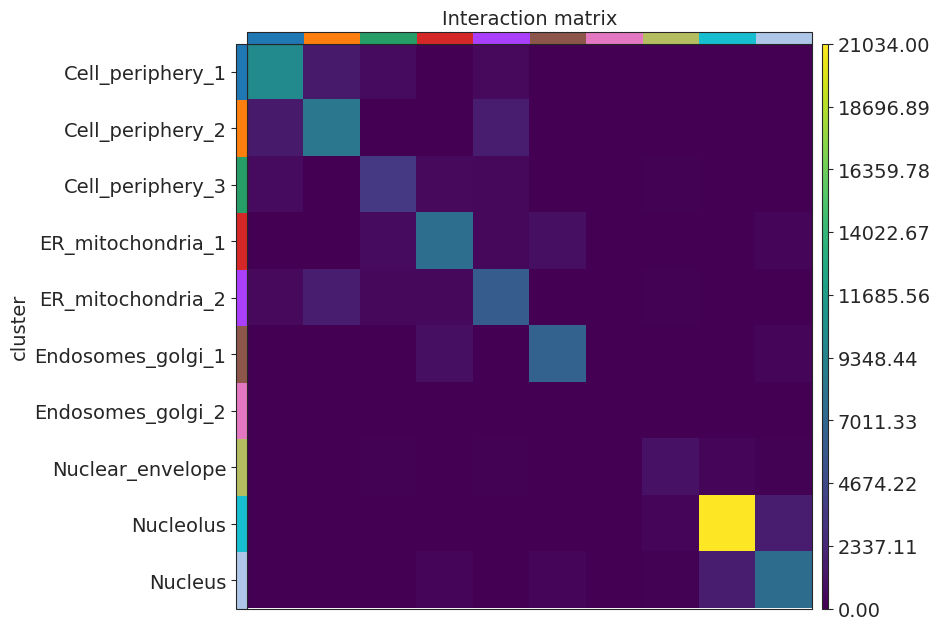
Interaction matrix and network centralities
[17]:
sq.gr.interaction_matrix(adata, cluster_key="cluster")
sq.pl.interaction_matrix(adata, cluster_key="cluster")
/home/yzy/anaconda3/envs/SODB/lib/python3.9/site-packages/squidpy-1.1.2-py3.9.egg/squidpy/pl/_graph.py:176: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
ad = AnnData(X=array, obs={cluster_key: pd.Categorical(adata.obs[cluster_key].cat.categories)})
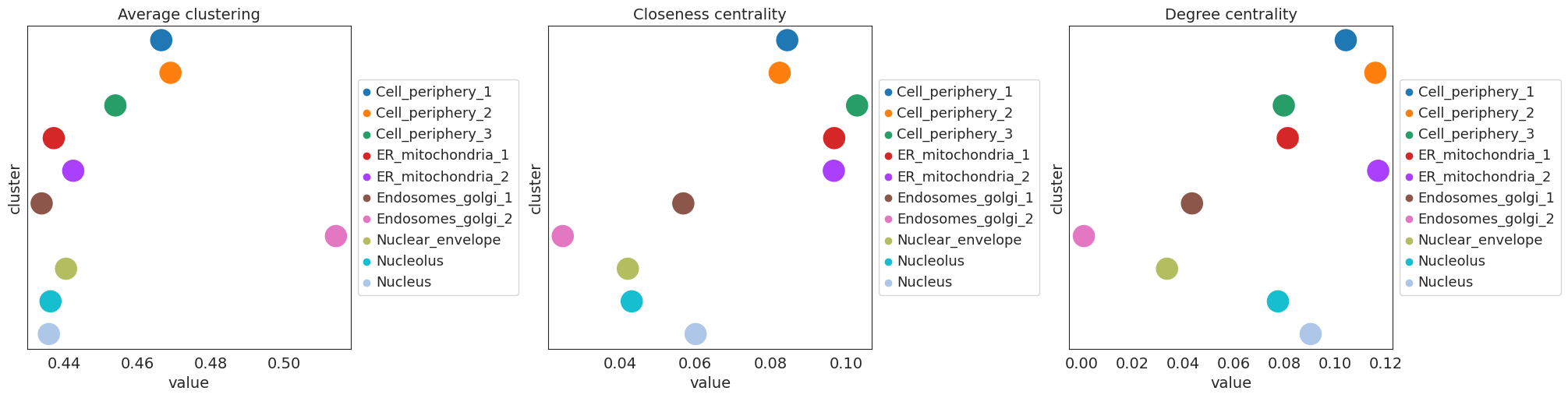
centrality score analysis
[18]:
sq.gr.centrality_scores(
adata,
cluster_key="cluster",
)
sq.pl.centrality_scores(adata, cluster_key="cluster", figsize=(20, 5), s=500)